Design the Model
Visit the Onyx website. Download Onyx, and create the pictured model.

LGC Model
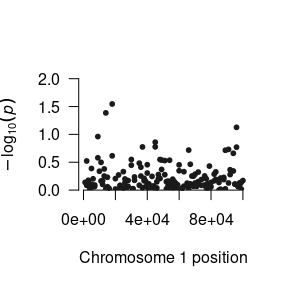
In this model, we have four occasions of measurement for the phenotype (t1, t2, t3, and t4). There are two latent variables, int for the intercept and slope for the slope of the trajectory. The loadings from int to the manifests are all fixed to 1. In contrast, the loadings from slope to the manifests increase. In OpenMx, it is possible to use the actual date of measurement instead of time increments of 1 unit. However, the use of non-unit time increments would force us to estimate the model using maximum likelihood, a method much slower than weighted least squares. So we recommend unit time increments. The single nucleotide polymorphism (SNP) data will appear in the snp indicator. It will be convienent to use exactly the name snp because gwsem knows which column to use by its name.
We export the model. This model does not entirely suit our purpose. After copying the code to a file, we remove the bits relating to modelData. We will also need to make a few changes to accomodate covariates. To make these changes easier, we gather up all the mxPath statements into a list.

model export
manifests<-c("t1","t2","t3","t4","snp")
latents<-c("int","slope")
path <- list(mxPath(from="int",to=c("t1","t2","t3","t4"), free=c(FALSE,FALSE,FALSE,FALSE), value=c(1.0,1.0,1.0,1.0) , arrows=1, label=c("int__t1","int__t2","int__t3","int__t4") ),
mxPath(from="slope",to=c("t1","t2","t3","t4"), free=c(FALSE,FALSE,FALSE,FALSE), value=c(0.0,1.0,2.0,3.0) , arrows=1, label=c("slope__t1","slope__t2","slope__t3","slope__t4") ),
mxPath(from="one",to=c("int","slope"), free=c(TRUE,TRUE), value=c(0.0,0.0) , arrows=1, label=c("const__int","const__slope") ),
mxPath(from="snp",to=c("slope","int"), free=c(TRUE,TRUE), value=c(1.0,0.0) , arrows=1, label=c("snp__slope","snp__int") ),
mxPath(from="int",to=c("int"), free=c(TRUE), value=c(1.0) , arrows=2, label=c("VAR_int") ),
mxPath(from="slope",to=c("slope","int"), free=c(TRUE,TRUE), value=c(1.0,0.1) , arrows=2, label=c("VAR_slope","COV_slope_int") ),
mxPath(from="t1",to=c("t1"), free=c(TRUE), value=c(1.0) , arrows=2, label=c("VAR_err") ),
mxPath(from="t2",to=c("t2"), free=c(TRUE), value=c(1.0) , arrows=2, label=c("VAR_err") ),
mxPath(from="t3",to=c("t3"), free=c(TRUE), value=c(1.0) , arrows=2, label=c("VAR_err") ),
mxPath(from="t4",to=c("t4"), free=c(TRUE), value=c(1.0) , arrows=2, label=c("VAR_err") ),
mxPath(from="snp",to=c("snp"), free=c(TRUE), value=c(1.0) , arrows=2, label=c("snp_res") ),
mxPath(from="one",to=c("t1","t2","t3","t4","snp"), free=F, value=0, arrows=1))
model <- mxModel("lgc",
type="RAM",
manifestVars = manifests,
latentVars = latents,
path)